Associate Professor Zheng Jie’s lab in SIST has recently proposed an advanced artificial intelligence model named KR4SL. By integrating knowledge graphs and explainable graph neural networks, KR4SL has made strides in identifying anti-cancer drug targets based on synthetic lethality. This work has been formally published as a Proceedings paper in the top international bioinformatics conference—the 31st Annual Conference on Intelligent Systems for Molecular Biology and the 22nd Annual European Conference on Computational Biology (ISMB/ECCB 2023). Furthermore, this work has been included in the ISMB/ECCB 2023 special issue of the Journal of Bioinformatics. Ms. Zhang Ke, a PhD student in Zheng’s lab, delivered an oral presentation at the conference. There were 336 manuscripts submitted to the ISMB/ECCB 2023 conference, with 60 selected as Proceedings papers, resulting in an acceptance rate of 17.9%.
Synthetic lethality (SL) is a type of mutual interaction between genes. Specifically, the inactivation of a single gene does not affect cell viability, while inhibiting two genes can result in cell death. Exploiting SL allows for the selective killing of cancer cells with mutated genes, while sparing normal cells. Even if certain mutated genes cannot directly serve as drug targets, cancer cells can still be attacked by targeting their SL partner genes. Current methods for identifying SL gene pairs include wet-lab experimental screening and computational prediction. Wet-lab experimental methods are typically expensive and may suffer off-target effects. Furthermore, the biological mechanisms behind many identified SL genes remain unclear. Knowledge graphs (KGs) encompass vast amounts of structured knowledge, which is able to support the computational prediction and interpretation of SL gene pairs. However, existing computational methods rely on the extraction of subgraphs from knowledge graphs to learn gene representations. These methods cannot fully utilize all the structural information available in the knowledge graphs, and it is also difficult to interpret them and make predictions.
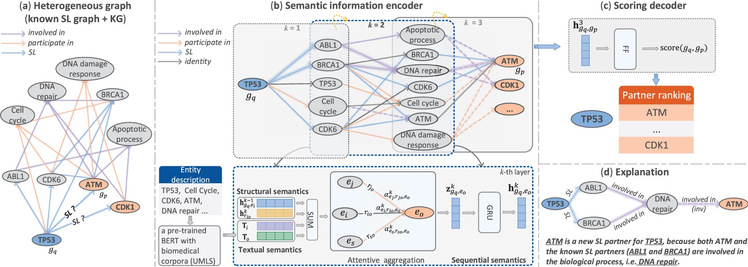
Figure: KR4SL model. (a) Integrate existing SL relationships and a knowledge graph to construct a new heterogeneous graph. (b) The diagram in the top row illustrates searching potential SL partner genes for a primary gene within a directed graph. The diagram in the bottom row shows the integration of three types of information and using attention mechanism to aggregate messages. (c) Generate scores for a candidate SL partner gene based on the representation of the gene pair. (d) Relational paths explain the prediction process and possible SL mechanisms
To address the limitations of existing computational methods in utilizing KGs, Zheng’s group proposed an explainable graph neural network model based on knowledge graph reasoning named KR4SL. Drawing inspiration from dynamic programming, KR4SL efficiently finds relational directed graphs for multiple gene pairs. It integrates structural information of the relational directed graph, sequential information of the relational paths, and textual information of entities. This approach facilitates a thorough understanding of the semantic representation of gene pairs to predict new SL interactions. Notably, KR4SL is an explainable AI model. Through weighted relational path-based reasoning, users can clearly trace the entire prediction process, understanding the rationale and logic behind the predictions. Extensive experimental results have demonstrated that KR4SL outperforms the state-of-the-art baseline methods and exhibits powerful generalization capabilities in two realistic scenarios.
Zheng’s group made the primary contribution to this study, and the work has been published in an article titled “KR4SL: knowledge graph reasoning for explainable prediction of synthetic lethality”. Zhang Ke is the first author. Other authors include PhD student Feng Yimiao in Zheng’s lab, Liu Yong, Senior Research Scientist from Nanyang Technological University, and Wu Min, Senior Scientist from the Agency for Science, Technology and Research (A*STAR), Singapore. Prof. Zheng Jie is the corresponding author.
*This article is provided by Prof. Zheng Jie




 沪公网安备 31011502006855号
沪公网安备 31011502006855号
